Heaven, they’re in heaven,
And their hearts beat so that they can hardly speak
And they seem to find the happiness they seek
When they’re in the membrane, signaling, cheek to cheek
The lyrics above, with apologies to Irving Berlin, describe protein-protein interactions on the cell membrane, most of which occur between “cheek to cheek” protein neighbors, or microenvironment protein neighbors. Although these neighbors, or signaling partners, have been in the proximity labeling spotlight, that spotlight has been too widely focused to isolate them from more distant proteins. Consequently, signaling partners on the cell membrane are often lost in the protein crowd, complicating the development of cancer medications and other therapeutics.
A more tightly focused spotlight has been developed by David W.C. MacMillan, PhD, the James S. McDonnell distinguished university professor of chemistry, and his team at Princeton University, in collaboration with scientists at the Merck Exploratory Science Center (MESC) in Cambridge. The new spotlight, which is called MicroMap (μMap), achieves uncommon spatiotemporal resolution. It is also highly selective. It can center on a target protein and illuminate only those proteins within a radius of 1 to 10 nanometers.

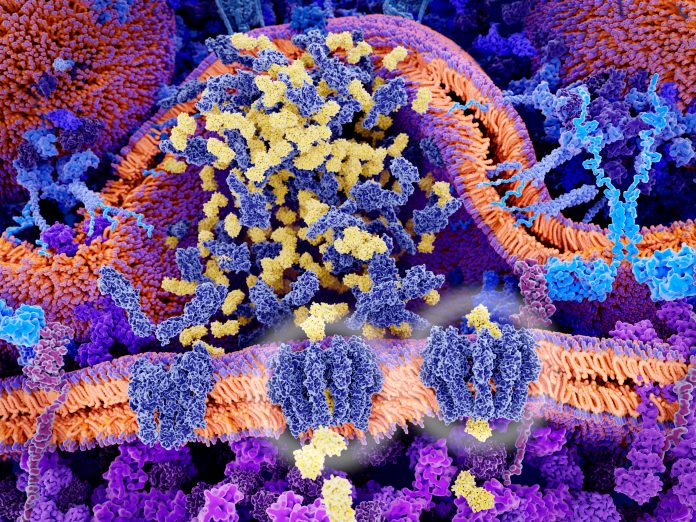
Details about μMap appeared March 6 in the journal Science, in an article titled, “Microenvironment mapping by means of Dexter energy transfer on immune cells.” The Dexter energy transfer is a short-range phenomenon, one that involves a catalyst that, upon illumination with blue light, transfers energy to a labeled probe molecule that incorporates diazirine. The catalyst can be engineered to selectively attach to any of the roughly 40,000 proteins on a cell’s surface. The probe, too, may be engineered to attach to a particular protein. When the diazirine in the probe is close enough to the catalyst to be energized, it breaks up, releasing carbenes, which readily attach to any proteins nearby.
“The [carbene] intermediates contain biotin, so the nearby proteins that they attach to become ‘painted’ with biotin,” explained Jacob B. Geri, PhD, a MacMillan group member and the Science article’s first author. “Using well established biochemical tools, those biotin-painted proteins are then identified using bottom-up mass spectroscopy.”
In this way, a target protein’s neighbors can be revealed by μMap, a technology that could impact proteomics, genomics, and neuroscience, to name a few of the more obvious fields. But the applications for fundamental biology are so wide-ranging that MacMillan is eager to get the technology “into everyone’s hands” to see what scientists in other fields can come up with.
MESC scientists and paper co-authors Rob Oslund, PhD, and Olugbeminiyi Fadeyi, PhD, said the technology could inspire vast new developments in biology. “Given the important role of understanding protein interactions within cellular microenvironments,” Oslund noted, “this technology has the potential to be a game-changing tool for both academic and industry life science labs all over the world.”
The scientists indicated that their catalyst accomplishes energy transfers repeatedly, triggering proximity-labeling reactions repeatedly, potentially labeling multiple localized molecules, proteins, and enzymes. Because carbenes are so short-lived—just a couple of nanoseconds—their reaction provides for a vivid, real-time snapshot of all contiguous molecules. Subsequently, researchers can quilt together a precise map of the microenvironment.
Besides mapping signaling partners on single cells, μMap may reveal “intrasynaptic” or “intersynaptic” interactions in two-cell systems. “A lot of the mechanism of disease takes place through how these cells talk to each other, and they can only talk if they’re touching,” explained Geri. “That’s why the surface of the cell is so important. If [two cells] touch, they can communicate.”
To demonstrate μMap’s ability to tap into cell-cell communication, the scientists used their technique to identify “the constituent proteins of the programmed-death ligand 1 (PD-L1) microenvironment in live lymphocytes and selectively labeled within an immunosynaptic junction.”
Normally, sick cells like cancer cells would present as molecular interlopers that need to be cleared by the immune system. But cancer cells are deceptive, said MacMillan. They send out a “don’t kill me” signal through a cloaking mechanism involving the PD-L1 and PD-1 axis. Since cancer therapies are successful partly based on their capacity to block that signal, scientists want to know more about how it is transmitted. Mapping the precise neighborhood is an essential early step.
“The μMap catalyst was put on antibodies that selectively bound to PD-L1 or PD-1,” detailed Geri. “Subsequent chemical events identified the proteins in proximity to PD-L1; proteins in proximity to the PD-L1/PD-1 complex are being investigated through ongoing studies.”
“Now, we don’t do the cancer biology,” added MacMillan. “But we’ve invented this tool that can give you a lot of information about these cancer cells. We think that by using this information, you can start to target those proteins as a way to also remove interfering signals. And if you can remove those signals, you make your immune system better at going after these cancer cells.”
“[Our] findings clearly demonstrate that the capacity of μMap to elucidate protein-protein interactions can be directly translated toward the highly selective labeling of dynamic interfaces within complex multicellular systems,” the article’s authors concluded. “We expect that this technology will find immediate use in antibody target identification, exploration of signal transduction pathways, profiling cell-cell junctions, and other disease-relevant microenvironments.”