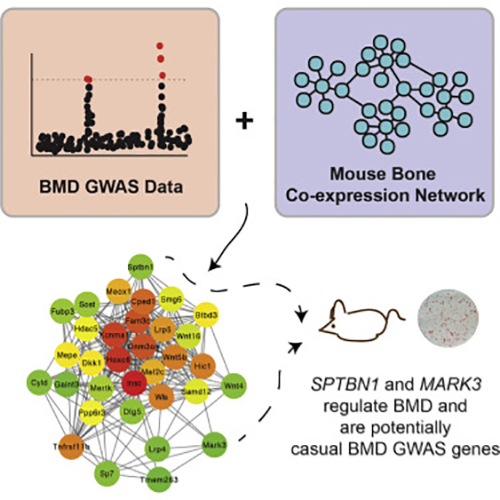
A new study led by researchers at the University of Virginia (UVA) School of Medicine's Center for Public Health Genomics just took an important step in the fight against osteoporosis. By combining data from genome-wide association studies (GWAS) onto a bone co-expression network, the researchers were able to predict and infer the function of a number of disease-associated loci. The findings from the study were published recently in Cell Systems through an article entitled “Integrating GWAS and Co-expression Network Data Identifies Bone Mineral Density Genes SPTBN1 and MARK3 and an Osteoblast Functional Module.”
Osteoporosis is a serious brittle bone disease that affects millions, and the UVA researchers identified more than a dozen genes amid the vast human genome likely responsible for bone density and strength. The new approach the researchers used to identify these genes could speed the development of new and better treatments for osteoporosis, as well as many other diseases.
In charting the genome, scientists commonly rely on GWAS, as these studies identify locations within the genome where genes associated with a certain disease are thought to be located. The problem, though, is that GWAS alone doesn't tell them which genes are truly influencing a disease.
“What's really challenging is going that next step and figuring out which genes are responsible,” explained senior study investigator Charles Farber, Ph.D., assistant professor at the University of Virginia School of Medicine's Center for Public Health Genomics. “It's similar to dropping a pin on a map app. So, the GWAS drops the pin, but it doesn't tell you anything about what's going on at that location, the mechanism through which genetic variants influence a disease.”
Dr. Farber and his colleagues wanted to go beyond GWAS, so they identified genes that appeared to work together and then mapped those genes onto the locations identified by GWAS. By cross-referencing the two, they were able to predict 33 genes that they believe are responsible for controlling bone density. Eighteen of the genes had been shown previously to play a role, but the remaining 15 were new.
“Many were genes known to have a role in the regulation of bone mineral density. In fact, over half of them were,” Dr. Farber noted. “So, that was a good proof of principle.”
Since their initial findings, the researchers tested two of the previously unknown genes and confirmed that they contribute to controlling bone mineral density. The UVA team does not expect their predictions will have a 100 percent success rate, but they do feel the technique has great promise for helping accelerate the process of determining gene function. Moreover, by hastening the understanding of gene function, they can accelerate the process of developing new drugs to target those genes, in order to treat disease.
“This was a way that we could take existing data and make predictions without going locus by locus without any direction,” Dr. Farber stated. “At least now we have hypotheses that we can test. I think that will speed up future attempts at trying to figure out which genes are truly causative.”











