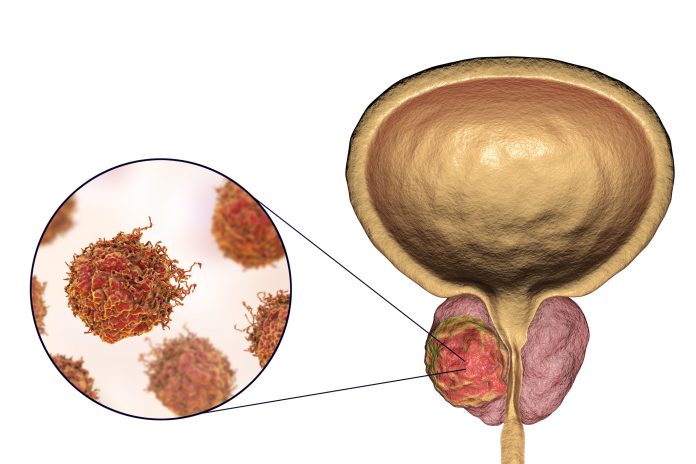
The molecular function of 87 inherited genetic variants affecting the risk of prostate cancer have been identified in a new study using three-dimensional mapping, and the majority appear to control the activity of genes located far away from the risk variants themselves. The study, which was published in Cancer Discovery, also found individual risk alleles could regulate multiple genes.
The approach this team used elucidates SNP function ad describes the germline-somatic interplay in transcription control. The researchers believe it can not only lead to discovery of new targets, but could help guide personalized treatment.
“Integrating 3D genome data with polygenic risk scores (PRS) can provide insights into genetic pathways associated with an individual’s lifetime prostate cancer risk and can usher in the development of intervention strategies to prevent or delay the disease,” said study leader Ram Mani, Ph.D., Assistant Professor of Pathology and Urology at UT Southwestern, and a member of the Harold C. Simmons Comprehensive Cancer Center.
Spatial transcriptomics, which reveals gene expression and its location, is one of the fastest growing fields in science.
“Our discoveries build on prior studies where 3D genome interaction data was used to resolve complex genetic puzzles, and, in so doing, provide a conceptual framework for transcription control by non-coding variants,” said Rami. He added that, “We suggest that 3D genome information is quintessential to deconvolute the relationship between the location of a risk SNP from its actual function.”
Hereditary prostate cancer has the highest heritability of any major cancer in men. So far, at least 185 common prostate cancer risk alleles have been found in families of European descent. But the vast majority of these alleles are located in DNA’s noncoding regions, so how they affect prostate cancer risk is largely unknown.
In their study, Mani and his colleagues used a 3D mapping technique and data from 565 prostate cancer tumors to show that 87 risk alleles affected the activity of hundreds of genes.
The cells being affected by these variants were mainly epithelial, but also included stromal cells and smooth muscle cells that support the epithelial cells. These genes produce proteins known to be involved in molecular pathways for development, apoptosis, and metabolism, among other cellular processes.
Some alleles had opposing activity on the multiple genes they affect. For example, one allele, known as rs8102476, simultaneously increased the activity of one gene while decreasing the activity of a neighboring gene. Risk alleles also had significant interaction with genes that acquired non-heritable mutations associated with prostate cancer. Of note, these variants appeared to predict how aggressive a patient’s disease became.
“Traditionally, we think of regulatory elements in the genome affecting neighboring genes,” said Mani. “But these risk variants, or risk alleles, can act like a light switch. The light is on the ceiling, but the switch is on the wall on the other side of the room.”
Together, Mani said, these findings could lead to better risk models for patients as well as new prostate cancer treatments. Further association studies are urgently needed to identify risk alleles in populations of non-European descent so that risk models and treatments can be customized to a broader patient base, he added.
“African American men have an elevated risk of prostate cancer, especially aggressive disease,” Mani said. “There’s a critical need for more genome wide association studies in this and other populations in which there’s an established health disparity to understand the nature of their risk and how to decrease it.”