Researchers at MIT and Harvard University have discovered a method of turning on gene expression only when a cell-specific or viral RNA is present, which should make RNA therapeutics safer and more effective in future.
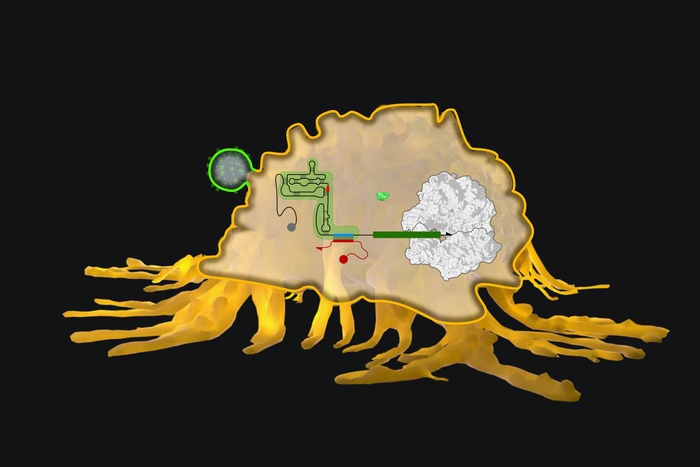
Dubbed ‘eToeholds’ these small RNA-based markers make use of internal ribosome entry sites (IRESs), which are small sections of RNA found in some viruses and eukaryotes and have evolved to start protein translation without relying on mRNA 5′ capping and polyadenylation.
“These are complicated folds of RNA that viruses have developed to hijack ribosomes because viruses need to find some way to express protein,” says Evan Zhao, a research fellow at the Wyss Institute for Biologically Inspired Engineering at Harvard University, who was one of the lead investigators involved in the study.
These IRES allow the eToeholds to allow expression of a linked protein-encoding gene sequence only when a specific cell or viral sequence is present. This control means that ‘off target’ effects, which have long been a worry for advanced therapeutics, can be largely eradicated making these kinds of therapies safer and more targeted.
“This brings new control circuitry to the emerging field of RNA therapeutics, opening up the next generation of RNA therapeutics that could be designed to only turn on in a cell-specific or tissue-specific way,” says James Collins, a professor at MIT and the senior author of the study, which is published in Nature Biotechnology.
Making sure that genetic therapies are only activated in disease causing cells and tissues has been challenging for researchers until now. “Current strategies are limited by low fold changes in transgene output and the size of trigger RNAs,” write the authors.
Collins and colleagues developed a similar system in bacterial cells a few years ago but wanted to transition the technology over to eukaryotes so it could be used to help design better therapies.
Using this new eToehold system they managed to detect a variety of different triggers in human and yeast cell lines in the lab. These included mRNA encoding Zika virus genes and also genes from SARS-CoV-2. They also detected proteins present naturally in human cells such as those that are produced in response to heat stress.
The researchers also think this system could help design better cancer therapies and one of the triggers they managed to detect was tyrosinase, an enzyme found in melanoma cells.
In terms of productivity, the team found that up to 16-fold production of the therapeutic proteins could be achieved using this method.
While this method has so far just been tested in the lab, the researchers believe it has a lot of potential to improve genetic medicine in both humans and other animals and are working on adapting it for further studies in animals before moving on to clinical trials.
“The ability to initiate translation of a desired protein in response to the presence of cell-type-specific or cell-state-specific RNA transcripts, as demonstrated here, has considerable therapeutic potential,” conclude the authors.
“We expect that future eToehold designs will implement logic gates to allow computation of cell states from multiple trigger RNAs, which might further enhance the specificity and utility of this system.”