The SARS-CoV-2 virus uses a specific ribosomal frameshift mechanism in order to reproduce inside host cells, something rarely seen in humans, and an Irish-Swiss research collaboration believes this could be a good target for possible therapies to fight the infection.
A programmed frameshifting process is used by all coronaviruses and is needed for viral RNA-dependent RNA polymerase to be produced and for the production of proteins needed for the virus to survive in its host.
However, during normal mammalian protein translation, frameshifting is very rare and if it occurs causes significant health problems due to dysfunctional protein production. For this reason, the researchers think this process could be targeted by medicines being developed to treat COVID-19.
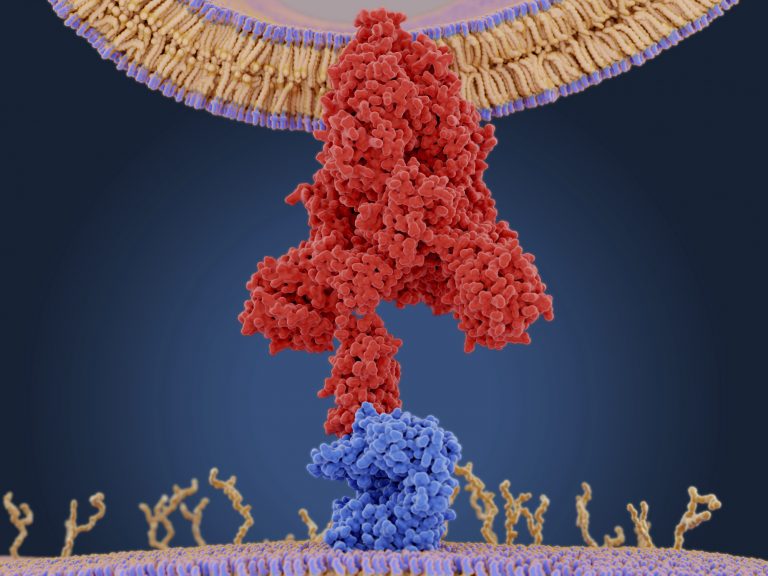
The research team used high quality cryo-electron microscopy to capture the process of mammalian translation of viral RNA in unprecedented detail. They discovered the virus uses a ‘pseudoknot’ structure that is able to create tension in the ribosome and promote frameshifting to translate viral RNA. They also observed that the young viral proteins seem to interact quite specifically with the ribosomal structure.
“The observation that the pseudoknot acts as an obstacle to slow down translation as the ribosome approaches the slippery site is mechanistically reasonable,” write the researchers in the journal Science.
“Since the pseudoknot is a stable structural element in the mRNA, it will resist unfolding and consequently generate a back-pull on the viral RNA, resulting in an increased chance of -1 frameshifting as the tRNAs are translocated.”
The team, led by John Atkins and Nenad Ban, both professors at University College Cork and ETH Zurich, respectively, confirmed their initial microscopy findings biochemically.
“Using computational modeling and reporter assays, compounds that have been predicted to bind the pseudoknot and inhibit SARS-CoV-2 frameshifting were described, but never tested with respect to their ability to inhibit viral replication,” explain the study authors.
To obtain proof of concept that SARS-CoV-2 frameshifting could be a good target for drugs to treat COVID-19, the scientists tested two of the compounds previously suggested to be good drug candidates on infected monkey cells.
Both compounds appeared to be non-toxic and reduced SARS-CoV-2 levels in the cells 3-4-fold. One of the compounds, merafloxacin, appears to be a better candidate for targeting frameshift changes as it showed a specific reduction in frameshifting relating to concentration. The other compound, MTDB, also seemed to reduce viral levels but through a different pathway.
This research is very early stage and these compounds are not currently potent enough to develop as possible drugs, but it provides a good starting point to develop new therapeutics for COVID-19. Such drugs might also have potential to treat other coronaviruses, as the frameshifting process is shared across all these viruses.
“Our future work will focus on understanding the cellular defense mechanisms that suppress viral frameshifting, as this could be useful for development of small compounds with similar activity,” says Ban.