A study led by Davide Marnetto, Ph.D., from the Institute of Genomics of the University of Tartu, Estonia and coordinated by Luca Pagani, Ph.D., from the same institution and from the University of Padova, Italy, has proposed a method to extend polygenic scores—the estimate of genetic risk factors and a cornerstone of the personalized medicine revolution—to individuals with multiple ancestral origins.
The researchers detailed their findings in the study “Ancestry deconvolution and partial polygenic score can improve susceptibility predictions in recently admixed individuals” published in Nature Communications.
“Polygenic Scores (PSs) describe the genetic component of an individual’s quantitative phenotype or their susceptibility to diseases with a genetic basis. Currently, PSs rely on population-dependent contributions of many associated alleles, with limited applicability to understudied populations and recently admixed individuals. Here we introduce a combination of local ancestry deconvolution and partial PS computation to account for the population-specific nature of the association signals in individuals with admixed ancestry,” write the investigators.
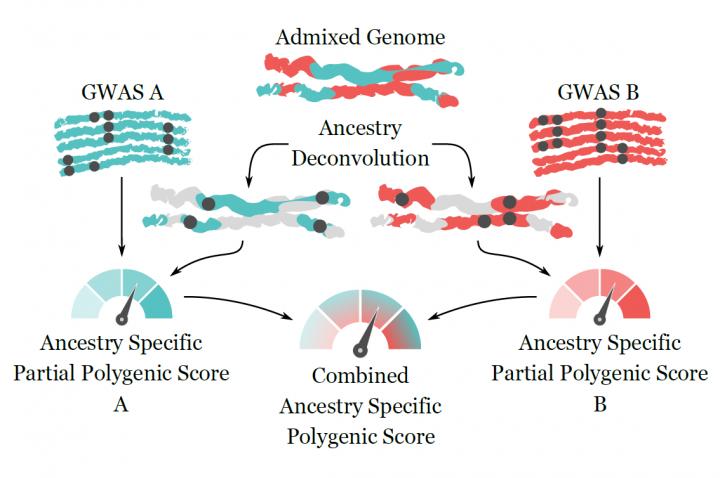
“We demonstrate partial PS to be a proxy for the total PS and that a portion of the genome is enough to improve susceptibility predictions for the traits we test. By combining partial PSs from different populations, we are able to improve trait predictability in admixed individuals with some European ancestry. These results may extend the applicability of PSs to subjects with a complex history of admixture, where current methods cannot be applied.”
“The information contained in our DNA is a mosaic of genetic instructions inherited from our ancestors, and in many societies one’s ancestors often come from the opposite corners of the world.” says Marnetto, first author of the study. The contribution of one’s ancestry, or ancestries, to the total risk of developing a specific disease or presenting a given trait is a long standing question of medical genomics. Nevertheless, most of the genotype/phenotype association data come from relatively uniform populations, in order to have a simplified and clearer picture. But what can be done when dealing with individuals who derive their ancestry from two or more distantly related populations?
“The latest developments of personalized medicine needed an extra step to be applied to individuals with more diverse origins, and here we tried to combine knowledge from homogeneous populations into a model that could work for recently admixed individuals” continues Marnetto.
To separate the various genomic components of each individual, Marnetto and colleagues applied methods from molecular anthropology and population genomics. “This research is a welcomed example of deep synergy between evolutionary/population genetics framework and medically oriented large scale genomics science, which is one of the focuses of our institute.” Adds Mait Metspalu, PhD, who is heading the institute of Genomics at the University of Tartu.
“Our work provides a solid proof of principle on the feasibility of using population genetic and molecular anthropology to boost the potential of personalized medicine. I hope our work can bring individuals of mixed ancestry one step closer to the benefits of personalized and predictive healthcare” says Pagani, the research coordinator.