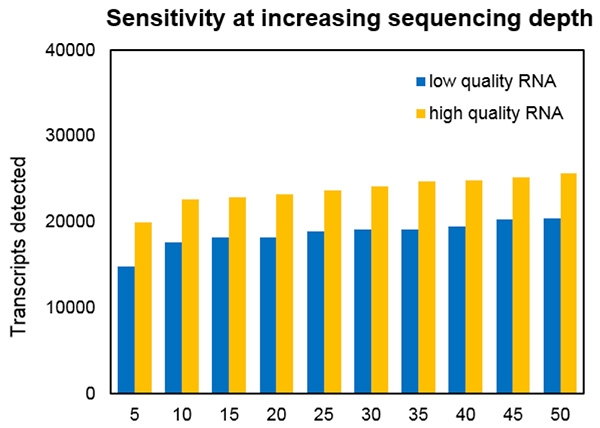
The greatest challenge to using RNA-Seq in the clinic arises from the massive amount of data produced—gigabytes of data from a single human transcriptome. Most RNA-Seq applications begin with aligning sequencing reads to the genome, a computationally intensive process requiring investment in hardware or access to a cloud-based platform. Post-alignment, downstream analysis is required to glean diagnostically relevant information, increasing the time to result. Consequently, RNA-Seq has the potential to provide a breadth of clinical information, but at the expense of the rapid response necessary for patient care.
Producing RNA-Seq Data
In a typical RNA-Seq experiment, RNA is first extracted from a cell culture or tissue sample. Many of these samples’ RNA molecules are ribosomal RNA, mitochondrial RNA, or other highly expressed transcripts, and so are not of clinical interest. Because of this, total RNA is often enriched for the relevant transcripts by poly-A pull-down or ribo-depletion; the latter process often involves noncoding transcripts. Enriched RNA then undergoes standard library construction, including reverse transcription, adapter ligation, and PCR.
At present, libraries are sequenced using high-throughput, short-read technology termed next-generation sequencing. Although Illumina platforms are commonly used, clinical applications of RNA-Seq, like transcript quantification and gene fusion discovery, can be accomplished using any short-read technology—presuming appropriate parameters are chosen.
To read the rest of this article click here.












