By Michael N. Liebman
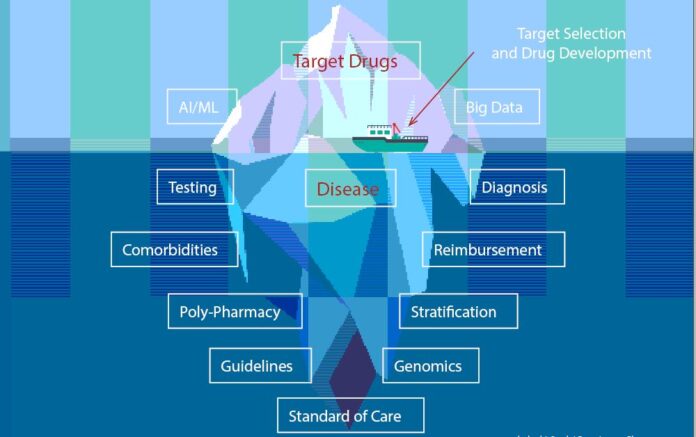
Biology embraces the existence of a central dogma, i.e., DNA → RNA → protein, and drug development has embraced its own, i.e., disease → target → drug, but both recognize the complexity that governs those transitions. The average cost of developing a drug approved for human use approaches $2 billion and takes 14 years on average, but also realizes a failure rate of approximately 90%. This limited rate of success suggests that significant challenges may exist at the system level, warranting the acquisition, testing, and incorporation of innovative approaches and technologies to address this disparity. Notably, the use of artificial intelligence (AI), machine learning (ML), and big data represent the latest wave of methods to be tested.
Computational approaches to the drug development process have long focused on the discovery phase, with an emphasis on target and drug selection and optimization. Historical emphasis on small molecule drug discovery, beyond direct chemical screening, led to the exploration of computational approaches like the pharmacophore model in the 1960s. This led to quantitative structure activity analysis; analysis of intermolecular complexes and molecular docking to three-dimensional protein structures (with advancements in molecular graphics); and molecular dynamics simulations. More recently, computational approaches have enabled the integration of genomic data with clinical records, claims data, and wearables; the simulation and computation of pK and pD values; and the prediction of protein three-dimensional structures from amino acid sequences. Downstream in drug development, AI methods are being applied to clinical trial design, patient enrollment, and digital twins.
Advances in and access to AI and ML approaches, including deep learning, have significantly impacted many aspects of drug discovery as noted above, enabling the analysis of more complex and larger quantities of data. The results of these analyses are promising as they attempt to address challenges in drug development. However, these methods remain primarily correlative and not causal. AI has been used to both identify a target and design a drug candidate, INSO18_055 (Insilico Medicine), which is now in clinical trials. But to date, there have been no FDA-approved drugs developed solely using AI methods. The goal is to use AI to reduce the failure rate in drug development and produce more effective therapeutics to improve patient care. But instead of asking whether AI will be used to design and develop drugs, it would be better to acknowledge that going forward, all drugs will be developed with the application of AI, whether in discovery or clinical trial development. In the 1970s, the question was “what drugs were developed by molecular modeling and molecular graphics?” The answer was “none, but all drugs are developed using these methods.”
While the results of these efforts, especially the application of AI and ML, are notable, they were focused on target selection and drug development rather than potentially critical aspects of diseases. The complexities of a disease and its accurate diagnosis are fundamental factors affecting root cause analysis of the current failure rate in drug development. Several key factors are outlined here and followed by clinical examples from triple-negative breast cancer and multiple sclerosis to highlight these complexities and their potential impact.
A re-focusing: Disease → Target → Drug
Although addressing the disease component of the drug development dogma may seem like an opportunity to reduce the current failure rate, it is not necessarily easy to accomplish by the computational or experimental approaches used for target identification and lead compound screening. Current estimates are that approximately 50–100 million (10%) of all conditions are misdiagnosed every year, resulting in 795,000 deaths and disabilities. This can significantly impact the accurate assignment of targets for drug development and undercut the need to stratify diseases into subtypes, which enables more personalized medicine. Among the challenges that need to be considered are:
- Disease is a process, not a state: Disease involves a set of processes that evolve over time and is not readily definable by a set of characteristics at a point in time.
- We do not know full untreated disease trajectories as it is unethical to not treat a patient upon diagnosis.
- Two patients may present the same lab values at a specific time point in their respective diseases but be on completely different disease paths.
- Two patients may present different lab values but be on the same disease path, just presented for diagnosis at different time points along their individual paths.
- A patient presenting at different stages of their disease can receive different diagnoses, resulting in different treatments and outcomes.
- A disease path requires specifying its vector dimension, i.e., based on essential clinical measurements collected over time; positionor progressionalong the disease path, i.e., disease staging; and rate of progression, i.e., velocity. Stage and speed of progression cannot be measured simultaneously (Heisenberg…).
- Disease stratification and phenotyping: Diagnosis and phenotyping are frequently used interchangeably, although “phenotype” is defined as an individual’s observable traits and is determined by both genomic makeup (genotype) and environmental factors.
- As noted above, failure to consider temporal progression of the disease during diagnosis can limit the potential to optimize treatment.
- Although all patients are unique at the highest resolution (n of 1), significant pragmatic improvement in patient management can be achieved through stratification into temporally similar subgroups, i.e., stratification or subtyping of the disease process itself.
- Stratified disease processes can reveal unique target opportunities for more successful drug development, including regulatory approval.
- Real-world evidence and data, claims data: AI and ML methods are most effective when accessing big data, which typically includes electronic health records (EHRs), claims data, and data from wearables. These data sources are developed and implemented for different purposes and the selection of data to be included in any AI/ML analysis should be driven by the question being addressed.
- Claims data reflects the focus on approval and reimbursement for services and are not an accurate representation of the underlying clinical presentation.
- The addition of “clinical notes” through natural language processing may be affected by use of “cut and paste” practices that have been introduced into EHRs.
- Data interoperability: All data are not equal, even if they have the same labels. Many efforts to create big datasets focus on interoperability across multiple databases to create virtual “patient cohorts.”
- Most efforts toward data aggregation focus on matching data fields, which frequently includes identifying equivalent data fields using semantic mapping.
- Data cleaning in these aggregation efforts typically adjusts for differences in units of measure and identifying potential “out of bound” values.
- EHRS are not specifically developed for research purposes and do not enable the critical level of annotation that should be included, although even if supported, most physicians would not have time for full annotation.
- Laboratory tests may only reflect “+” or “-” assessments.
- Data fields may reflect results obtained using different laboratory tests, e.g., antibodies.
- Data fields may represent differences in assigning thresholds to determine “+” vs “-” test results.
- Reproducibility may be operator dependent, e.g., histopathology.
- Algorithmically derived data fields may use different algorithms, e.g., eGFR evaluation involves one of five algorithms, two of which include race-based adjustments.
- Laboratory tests may only reflect “+” or “-” assessments.
- Pathways and processes: The genome partly functions as a major “parts list” for biological processes that rely on molecular pathways, serving as an underlying road map. Pathways are commonly viewed in terms of their component intermolecular interactions, as drawn in two-dimensions to minimize the overlap of components.
- Pathways are a convenient representation of many complex intermolecular interactions, but unlike in prokaryotes, those in eukaryotes do not appear to be coordinated products of genetic origin.
- Pathways function biologically in four-dimensional space, i.e., they have time dependency, as not all components may be present concurrently.
- Pathway models may be inaccurate or incomplete, either missing either critical elements and nodes or interactions, i.e., feedback loops, that are critical for producing the observed biological function.
- The simulation of pathway dynamics frequently uses individual reaction kinetic data measured under different experimental conditions and not necessarily suitable for incorporation into a model of system behavior.
- Linear pathways involve fewer nodes and while potentially more efficient, branching in pathways reduces the potential lethality of “breaking a link” and enables rapid adaptation and greater control of pathway behavior.
- Defining a real-world patient: A real-world patient is not well-represented in their EHR, but that is whom a physician must diagnose and treat.
- Genomics contains critical information related to potential function and dysfunction, i.e., the interaction with external factors determines potential responses and the manifestation of conditions or diseases. For instance, BRCA1 mutations significantly raise breast cancer risk but do not guarantee disease.
- Environmental exposure is not only dependent on “what” and “how much,” but also on “when,” especially during development or some chronic conditions that occur during gestation.
- Similarly, lifestyle is not only dependent on “what” and “how much,” but “when” as well, especially during development or some chronic conditions. For instance, it is not only “do you smoke and how much?” but “when did you start and how did it progress?”
- Clinical history: Real-world patients average 4–5 comorbid conditions, i.e., previously treated, currently being treated, and not yet diagnosed, which can lead to 8–10 medications including those that are prescribed, over-the-counter, and dietary or supplements.
- Social determinants of health: Increasing awareness of external factors that correlate and may cause chronic conditions, i.e., comorbidities.
- Cultural and trust determinants of health: As efforts are expanded to enhance the diversity of patient populations in clinical trials and drug development, it is important to consider that cultural and trust factors may not align with efforts to address what is being defined within social determinants of health.
- Defining real-world diversity: There is a significant difference between the patient in a clinical trial (or their digital twin) and the patient that a physician encounters in the office.
- Current efforts to enhance diversity focus on age, gender, ethnicity, and more recently, disability. There is increased pressure to include children and pregnant women (who were excluded from the initial COVID-19 vaccine trials).
- As noted above, real-world patients also have an extensive clinical history with comorbidities, experience polypharmacy, are exposed to external factors included in social determinants of health, modulate their responses based on commercial determinants of health and TDOH, are likely not aware of environmental or dietary exposures, and exhibit a wide-range of lifestyles, only some of which are discussed with the physician.
- While it is not practical to include the real-world spectrum of patients in a clinical trial, it might be possible to establish the characteristics of the patient population likely to assist physicians in understanding the gap between trial participants and the regulated label for an approved drug.
- Clinical guidelines and protocols: Clinical guidelines are commonly the result of the efforts of groups of practitioners and may differ significantly across specialties and international boundaries. Protocols commonly represent the implementation of guidelines that are adjusted for local conditions and preferences.
- Clinical guidelines, even when evidence-based, commonly reflect a consensus derived from the experiences of multiple experts who evaluate the level (confidence) of the evidence provided for each step in the guideline.
- Guidelines are separately developed but potentially coordinated for diagnosis and treatment. They undergo revision in a non-periodic manner to reflect new observational data, new technologies, and regulatory approvals.
- When developing cohorts from existing databases, it must be considered that patients diagnosed similarly may have been diagnosed using different guidelines. This extends to the use of ICD-10 codes.
- Protocols reflect the real-world implementation of guidelines that consider local resources, medical center policies, experience of the chief of service, and more recently, local politics.
These challenges act both independently and interdependently to establish the complexity that needs to be addressed in drug development and varies across therapeutic areas. Using systems-based analysis to expand the perspective, some key real-world challenges are described below for two complex conditions: triple-negative breast cancer and multiple sclerosis.
- Triple-negative breast cancer
Triple-negative breast cancer (TNBC) accounts for 10–15% of all breast cancers. The name refers to the fact that the cancer cells do not have estrogen or progesterone receptors (ER or PR) and make either little or no HER2 protein (i.e., the cells test “negative” on all three tests.) These cancers tend to be more common in women younger than 40 years, who are Black, or who have a BRCA1 mutation.- Patient management is largely based on ER, PR, and HER2 receptor levels, i.e., diagnosis, stratification, and treatment.
- Immuno-histochemistry (IHC)
- Measures anti-body response
- inter-laboratory variability
- Unsuccessful standardization of laboratory procedures
- Fluorescent in situ hybridization (FISH)
- Measures gene copy number in cells
- Significant reproducibility
- Overall reproducibility
- False positive rate: 14–16%
- False negative rate: 18–23%
- Discordance between IHC and FISH approximately 20%, grade dependent
- Higher in +1s and +2s
- IHC and FISH do not measure the same biological entity. The former measures protein level and the latter measures gene copy number.
- “Positivity” thresholds range from 1% to 10% or higher.
- Inter-lab/inter-pathologist variability is notable. For instance, analysis of positivity rates in 39 laboratories (33,794 inflammatory breast cancer patients) revealed 35.9% (ER), 43.6% (PR), and 28.2% (HER2) outside the 95% confidence level
- Intratumor heterogeneity is notable.
- HER2 levels can flip, changing from positive to negative or vice versa over the course of the disease.
- Clinical guidelines and threshold levels are typically not standardized, although some countries do have national standards.
- Approximately 50% of patients receiving trastuzumab have never been tested for HER2.
- Approximately 60% of patients receiving trastuzumab do not respond to the drug.
- No ICD code exists specifically for TNBC.
- Multiple current clinical trials (clinicaltrials.gov) use “TNBC diagnosis” for inclusion in trial.
- Immuno-histochemistry (IHC)
- Patient management is largely based on ER, PR, and HER2 receptor levels, i.e., diagnosis, stratification, and treatment.
- Multiple sclerosis
Multiple sclerosis (MS) is a degenerative disease in which insulating covers of nerve cells, called myelin sheaths, in the brain and spinal cord are damaged. This damage disrupts the ability of parts of the nervous system to transmit signals, resulting in a range of signs and symptoms like physical, mental, and sometimes psychiatric problems. Specific symptoms are double vision, visual loss, muscle weakness, and trouble with sensation or coordination. MS takes several forms, with new symptoms either occurring in isolated attacks (relapsing) or building up over time (progressive). In the relapsing forms of MS, symptoms may disappear completely between attacks, although some permanent neurological problems often remain as the disease advances.- Commercial: U.S. cost $28 billion/year ($19 billion in drug costs alone).
- Public health: > 1,000,000 cases in U.S. and 2,500,000 cases globally.
- Female:male ratio is about 3:1
- Approximately twice as prevalent above the 37th parallel (north vs south in the U.S. and cold vs warm climates)
- Low risk in Native Americans, African Americans, Africans, and Asians
- High clinical trial failure rate (Phase III).
- For drugs receiving FDA approval, low clinical acceptance rate.
- Average patient cost is $45,000/year.
- Misdiagnosis and missed diagnosis are both common and critical (1 in 5 diagnoses).
- No specific diagnostic test (MRI white matter correlative is not causal)
- Differential diagnosis requires ruling out migraine; Lyme disease; conversion disorders; neuromyelitis optica; lupus; stroke; fibromyalgia Sjögren’s syndrome; vasculitis; myasthenia gravis; sarcoidosis; vitamin B12 deficiency; and acute disseminated encephalomyelitis
- Average time to diagnosis is 5 years. During this period the patient is usually treated for symptoms, which may impact final diagnosis and response to therapy.
- Clinical guidelines for treatment continue to vary over time.
- Use of the Expanded Disability Status Scale (EDSS) score is biased towards walking or mobility
- Involves subjective evaluation of seven systems with potential degeneracy, as there may be heterogeneity among patients exhibiting the same EDSS score
(Note: Many guidelines include the use of “scores” to assist clinical diagnosis, but these can obscure the heterogeneity of real-world patients.)
- 85% of initial diagnoses are Relapsing Remitting MS (RRMS), with at least 3 subtypes.
- 10-year progression to secondary progressive MS (SPMS) (50%)
- 25-year progression to SPMS (40%)
- No progression (10%)
- Lifetime cost estimated at $4,000,000 per patient.
- Full economic burden is approximately twice the medical costs.
Challenges lead to opportunities for AI to improve drug development
Applications of AI/ML to drug discovery are beginning to produce results that can positively impact the current drug development process. However, success will be measured not only in terms of reducing the time needed to identify potential clinical candidates, recruit for clinical trials, and create digital twins, but ultimately in conducting successful clinical trials, gaining regulatory approval, and becoming commercially viable. One challenge is that we are attempting to use these new technologies to address what we perceive to be critical questions and are not considering that the questions need to evolve. Drug development has itself progressed from a chemistry (or natural product)-focused approach to disease or symptom management. However, this has not addressed the full complexity of a patient, clinical practice, or the disease process. The opportunity to significantly impact drug development will require dealing with the complexity of understanding the disease and patient population.
It is critical to recognize that disease is a process, not a state, and that disease stratification based on temporal clinical measurements and not claims data alone is essential to develop disease trajectories. The available data will be incomplete, contain errors, and be inadequately annotated to highlight differences in measurement methods and interpretative analytics. Clinical guidelines contain consensus-generated and increasingly evidence-based workflows for diagnosis and/or treatment, but are constantly evolving and can affect the diagnosis (and the diagnosis code) of a patient. These factors suggest that improving drug discovery by addressing the disease component will not be simple but can be crucial for improving the current failure rate in drug development. Critically, it can impact the outcome for both physicians and patients.
The optimal return on investment for implementing methods like AI/ML will be realized when they are used to address the questions that remain as “unknown unknowns.” Applying evolving technologies to “look for our keys under the lamppost” may make the search go faster, but not necessarily find them more than 10% of the time.
Michael N. Liebman, Ph.D is the Managing Director of IPQ Analytics, LLC and Strategic Medicine, Inc after serving as the Executive Director of the Windber Research Institute (now Chan Soon-Shiong Institute for Molecular Medicine) from 2003-2007. He is an Adjunct Professor of Pharmacology and Physiology at Drexel College of Medicine, Research Professor of Biology at University of Massachusetts-Lowell and Adjunct Professor of Drug Discovery, First Hospital of Wenzhou Medical University and also Fudan University.