Research led by Johns Hopkins University and Icahn School of Medicine at Mount Sinai has revealed a specific gene expression signature that differentiates people with long-term Lyme disease from those with acute disease or uninfected controls.
The scientists also identified a diagnostic signature including 35 genes for any type of Lyme disease, which they hope could help improve diagnosis for this infection in the future.
“We wanted to understand whether there is a specific immune response that can be detected in the blood of patients with long-term Lyme disease to develop better diagnostics for this debilitating disease. There still remains a critical unmet need, as this disease so often goes undiagnosed or misdiagnosed,” said Avi Ma’ayan, a professor at Icahn Mount Sinai, and senior author of the paper, in a press statement. “Not enough is understood about the molecular mechanisms of long-term Lyme disease.”
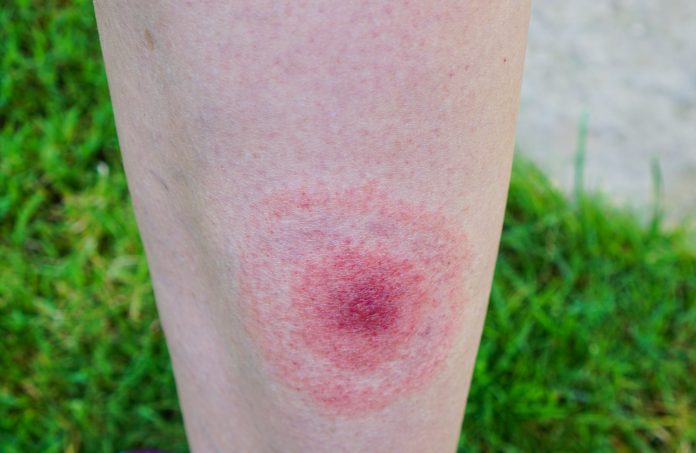
Lyme disease spread by tick bites and in most cases can be treated with antibiotics if caught early, but 10-20% of cases progress to a more chronic form of the condition. Numbers of infections are known to be on the increase, with 30,000 diagnosed cases reported to the CDC in the U.S. each year. However, the estimated real number is thought to be closer to 476,000 cases.
To try and assess molecular differences between chronic and acute cases of the infection, Ma’ayan and colleagues recruited 152 individuals with continuing, chronic symptoms of Lyme disease after treatment (PTLD), as well as 72 individuals with acute Lyme disease (LD) and 44 uninfected controls for comparison purposes.
The research team profiled the gene expression profile of the peripheral blood mononuclear cells of the participants with RNA sequencing and used machine learning to identify the most relevant genes.
As reported in in Cell Reports Medicine, results showed that most people with PTLD had a gene expression profile that was distinct from individuals with LD and healthy controls, although a small number of those with PTLD had profiles overlapping with LD cases.
The team also identified 35 genes expressed differently in LD and PTLD versus healthy controls. They think these proteins could help with Lyme disease diagnosis if whole blood could be analysed instead of peripheral blood mononuclear cells, as isolation of the latter is costly and requires academic laboratory expertise.
“Not enough is understood about the molecular mechanisms of long-term Lyme disease,” commented Ma’ayan. “We should not underestimate the value of using omics technologies, including transcriptomics, to measure RNA levels to detect the presence of many complex diseases, like Lyme disease. A diagnostic for Lyme disease may not be a panacea but could represent meaningful progress toward a more reliable diagnosis and, as a result, potentially better management of this disease.”