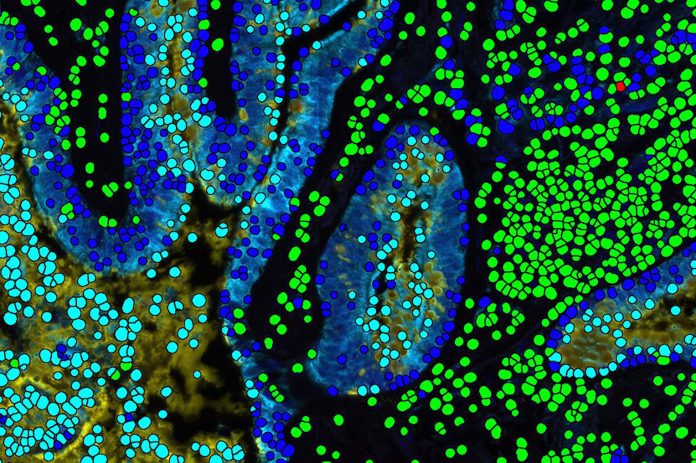
Researchers in the Mahmood lab at Brigham and Women’s Hospital have developed a new deep learning algorithm that is capable of teaching itself to search large datasets of pathology images to identify similar cancer cases. The tool, called SISH for “Self-Supervised Image Search for Histology,” has the ability to identify analogous features in pathology images and uses that information to both pinpoint the form of disease, while also helping doctors and other clinicians determine which therapies will be most effective for each patient.
Details of the algorithm were published today in the journal Nature Biomedical Engineering.
“We show that our system can assist with the diagnosis of rare diseases and find cases with similar morphologic patterns without the need for manual annotations, and large datasets for supervised training,” said senior author Faisal Mahmood, PhD, in the Brigham’s Department of Pathology. “This system has the potential to improve pathology training, disease subtyping, tumor identification, and rare morphology identification.”
Over the past 10 or more years, health systems have increasingly digitized the storage of tumor samples via whole-slide imaging (WSI) systems. While digital pathology databases can easily store the gigapixel sized images created by WSI, the sheer volume of data has made querying these data difficult.
“As institutions scan and store an increasing number of images, they often turn to WSI storage and retrieval paradigms identical to that used for their glass slides—large repositories of data searchable through patient identifiers, case number, date of procedure, pathology report and so on, without leveraging the digital morphologic content of the images themselves,” the researchers wrote. “A critical challenge that hinders large scale, efficient adoption of histology whole-slide image search and retrieval systems is scalability. This is a unique challenge for WSI retrieval systems as compared with other image databases since they need to efficiently search a growing number of slides that can each consist of billions of pixels and be several gigabytes in size.”
One factor that hinders the efficient search of WSI data lies in the fact the that majority of computational pathology methods currently used leverage supervised deep learning that rely on using slide or case level labels to solve classification or ranking problems. The investigators theorized that a self-teaching image search tool such as SISH, that harnesses the rich, spatially resolved information in pathology images, is much more powerful. Applications for such a tool include identifying cases with similar morphological features to assist in diagnosing rare cancers that might otherwise remain undiagnosed due to the lack of enough cases for accurate supervised classification models to be developed; or for finding cases with similar morphologies to predict outcome for clinical trials with limited samples.
For the Brigham and Women’s study, the researchers tested the speed and ability of SISH to retrieve interpretable disease subtype information for both common and rare cancers. The algorithm successfully retrieved images quickly and accurately from a database of whole slide images from over 22,000 patient cases, representing more than 50 different disease types from over a dozen anatomical sites. The speed of retrieval outperformed other methods in many scenarios, including disease subtype retrieval, particularly as the image database size scaled into the thousands of images. Even among larger repositories, SISH maintained a constant search speed.
The investigators did note some limitations of the technology, including a large memory requirements, limited context awareness within large tissue slides, the technology being limited to a single imaging modality.
However, SISH did demonstrate it can efficiently retrieve analogous images regardless of the size of the repository being queried, a refined ability to diagnose rare disease types, and an ability to serve as a search engine to recognize regions of images that may be relevant for diagnosis. In all, the investigators noted, SISH may help to inform future disease diagnosis, prognosis, and analysis.
“As the sizes of image databases continue to grow, we hope that SISH will be useful in making identification of diseases easier,” Mahmood concluded. “We believe one important future direction in this area is multimodal case retrieval which involves jointly using pathology, radiology, genomic and electronic medical record data to find similar patient cases.”